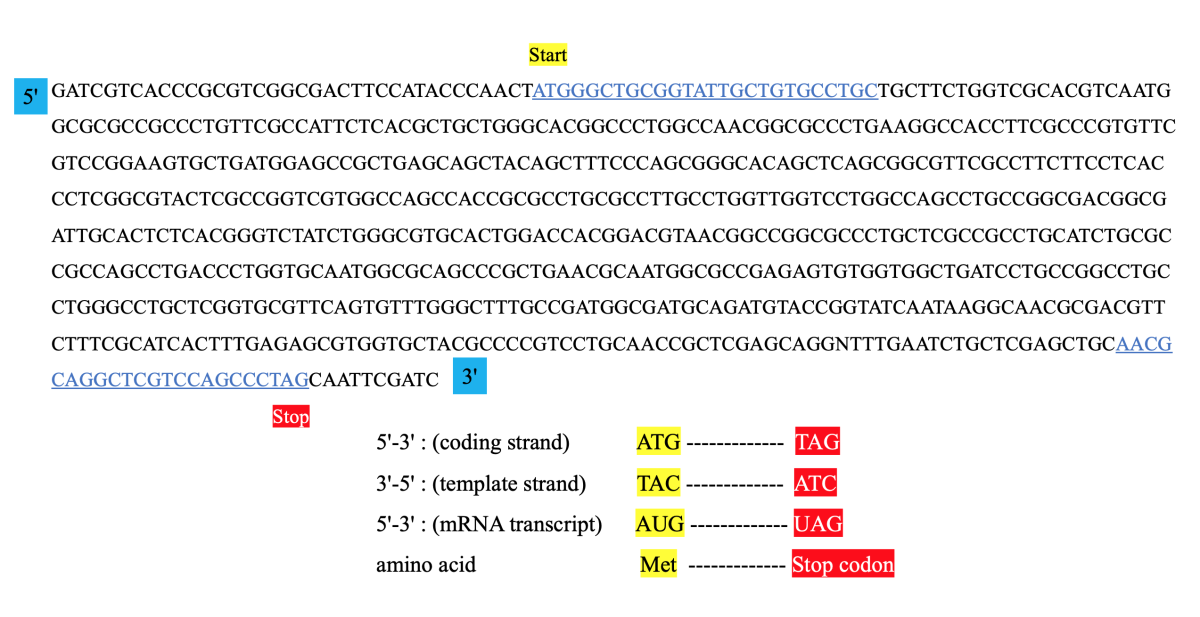
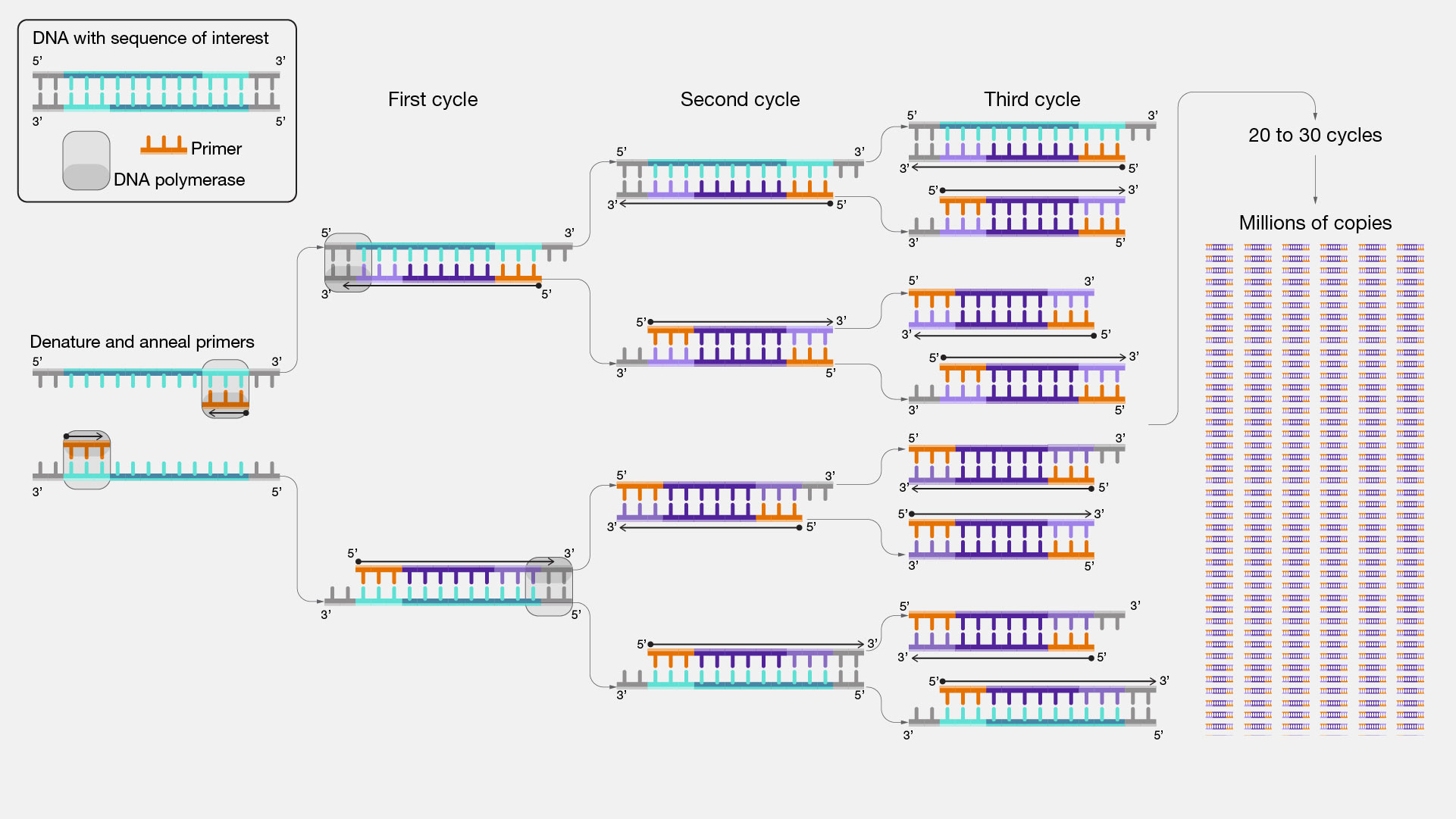
Design PCR primers and check them for specificity
Table Of Content
The use of DNA primers in PCR enables the targeted amplification of specific DNA regions for further analysis, such as DNA sequencing or detection of specific genetic markers. By designing primers that specifically bind to the desired target sequence, researchers can selectively amplify and analyze specific regions of the genome. Steps 4 and 5 are repeated until either a pre-determined generation g, or until L(Sg) is below a pre-determined threshold Lt. In our implementation, we typically run the optimization to about 1.5 × gt to ensure we reach local minima. To further improve the overall quality of the generated primer set, we recommend running multiple SADDLE optimization processes with different starting conditions (initial primer sets) and selecting the best final primer set.
NEBuilder HiFi DNA Assembly & Gibson Assembly
Multiplex qPCR thus typically requires significant empirical optimization, even at around 4-plex11. SADDLE allowed us to successfully design a 60-primer qPCR panel targeting 56 gene fusions, and exact fusion identities can be determined through affordable Sanger sequencing. Thus, we envision that SADDLE can revolutionize the use of qPCR for highly multiplex molecular diagnostics.
Primer Design using Gibson Cloning Method
Highly multiplexed rapid DNA detection with single-nucleotide specificity via convective PCR in a portable device - Nature.com
Highly multiplexed rapid DNA detection with single-nucleotide specificity via convective PCR in a portable device.
Posted: Thu, 01 Jul 2021 07:00:00 GMT [source]
The user can also draw new primers directly onto the canvas (Fig. 6, circle 4) which will return a JavaScript popup box with the DNA sequence, GC% and primer Tm values of the sequence that corresponds to the line drawn by the user (Fig. 6, circle 5). If the line is drawn from left to right, a forward primer is returned and if the line is drawn from right to left, a reverse complemented primer is returned in the popup box. Similar to section 1 above, the data can be uploaded locally in rs_fastA format (Fig. 2, circle 5) or retrieved remotely (Fig. 2, circle 6) from NCBI’s dbSNP23. SNP sequence data is preprocessed by PrimerMapper to collect the SNP position and type from the header field. Next, the primer design criteria are populated within the “PRIMER DESIGN” frame (Fig. 2, circle 7), followed by program execution after clicking buttons 1 to 3 within the “RUN” frame (Fig. 2, circle 8).
How can primer specificity be ensured?
Melting temperature (52°C-56°C) The GC results of the sequence gives a fair indication of the primer Tm. Solely living organisms utilize RNA primers while invitro involves DNA primers. However, DNA primers are much preferred due to varied reasons such as stability, easy storage, fewer enzymes required to initiate synthesis. Automatic genome annotation, real-time sequence analysis and protein structure prediction. Adding products to your cart without being signed in will result in a loss of your cart when you do sign in or leave the site.
The user can also click the ‘User Guide’ button to get an overview of each feature. (a,b) A sample graphical output generated by PrimerMapper for DNA based sequences in fastA format is shown. The primers designed to each sequence (in this case, two sequences), is converted to a graphical output with each primer represented by a blue glyph arrowed in the direction of synthesis. (c) PrimerMapper also generates a concatenated graphic that assembles each sequence (adopting the order followed in the inputted file) and its designed primers into a single view so as to quickly and easily validate the distribution of primers across all sequences. The amounts of these three species in the three primer set libraries are shown in Fig.
You can view Archer assays alongside IDT’s xGen™ NGS portfolio to find the best next generation sequencing solution for your lab. Many of the Swift products you have grown to love are now part of our new complete portfolio, xGen™ NGS. Through this new partnership we are pleased to offer you comprehensive next generation sequencing solutions. Apart from the PCR, DNA sequencing primers combine with restriction cloning, as well as other DNA new assemblies such as Gibson DNA assembly methods together with Golden Gate method. Designing oligonucleotides and making sure that you have the right parameters for your oligo is an important step in securing results, especially in PCR Primer Design.
We randomly selected a primer pair candidate for every amplicon that we wish to design, and collectively the selected primers are known as the initial primer set S0. This method demands the use of a vector assembly (plasmid) into a single construct with one or multiple DNA fragments. The PCR primers overlap to form restriction sites with adjacent DNA fragments and are designed, however, Type 2S enzyme, along with DNA ligases of the fragments for a directional assembly. Likewise, this method exploits the use of type 2 class of restriction sites, i.e. cut outside of their restriction sites through non-palindromic sticky end overhangs. This method exploits multiple fragments of DNA by using a combination of overhang sequences on their insert fragments for easy annealing with the adjacent ones.
Gene fusion detection with qPCR and sanger sequencing
This is not consistent with our understanding of DNA hybridization and polymerase extension kinetics, and implies that we may not be able to generate a perfect Badness function even ignoring computational resource constraints. This tool allows users submit sequence or multiple sequences in fasta format and return the best pair of primers near the sequence ends. Proper primer design is one of the single most important factors in successfulautomated Sanger DNA Sequencing.
This enables our new graphic display that offers enhanced overview for your template and primers. This specifies the range of total intron length on the corresponding genomic DNA that would separate the forward and revervse primers. The Tm calculation is controlled by Table of thermodynamic parameters and Salt correction formula (under advanced parameters). The default Table of thermodynamic parameters is "SantaLucia 1998" and the default Salt correction formula is "SantaLucia 1998" as recommended by primer3 program. Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Temporary primer set T is thus generated by combining this new primer pair with all remaining primers in set Sg. Optionally, multiple primer pairs can be replaced simultaneously in this step to allow faster and more efficient exploration of the primer set space. In our preliminary in silico evaluations, we found that simultaneously mutating multiple primer pairs generally caused a slowdown of the optimization process. For discovery applications, “shotgun” whole genome sequencing (WGS) is the preferred approach to identify novel DNA sequences of interest5. For routine detection of disease-relevant DNA variants in known genes of interest, targeted sequencing or direct qPCR approaches are typically used6,7.
Synthetic DNA templates were purchased as desalted DNA oligonucleotides (gBlocks, Integrated DNA Technologies), and stored at −20 °C. Human cell-line gDNA sample NA18562 (Coriell Biorepository) was stored at −20 °C. The gDNA was mixed with synthetic DNA templates at various ratios to create samples containing different proportions of a specific variant sequence. Dilution of gDNA samples and synthetic DNA templates were made in 1× TE buffer with 0.1% Tween 20 (Sigma Aldrich). At Takara Bio, we thoughtfully develop best-in-class products to tackle your most challenging research problems, and have an expert team of technical support professionals to help you along the way, all at superior value. Learn how to use our online tool to quickly and easily design primers for every In-Fusion Cloning project.
You can also lower the E value (see advanced parameters) in such case to speed up the search as the high default E value is not necessary for detecting targets with few mismatches to primers. A sample graphical output generated by PrimerMapper for browser visualization is shown. A canvas text file for each sequence is generated by PrimerMapper and these files can then be opened directly in a browser by clicking the “primermapper.HTML” file and loading the data. A sample configured text file is included in the download called “PrimerMapper_Sample.txt” which can be directly loaded into “primermapper.HTML” (circle 1).
In eukaryotes the removal of RNA primers in the lagging strand is essential for the completion of replication. Thus, as the lagging strand being synthesized by DNA polymerase δ in 5′→3′ direction, Okazaki fragments are formed, which are discontinuous strands of DNA. Then, when the DNA polymerase reaches to the 5’ end of the RNA primer from the previous Okazaki fragment, it displaces the 5′ end of the primer into a single-stranded RNA flap which is removed by nuclease cleavage. Cleavage of the RNA flaps involves three methods of primer removal.[4] The first possibility of primer removal is by creating a short flap that is directly removed by flap structure-specific endonuclease 1 (FEN-1), which cleaves the 5’ overhanging flap. This method is known as the short flap pathway of RNA primer removal.[5] The second way to cleave a RNA primer is by degrading the RNA strand using a RNase, in eukaryotes it’s known as the RNase H2. This enzyme degrades most of the annealed RNA primer, except the nucleotides close to the 5’ end of the primer.
Set a lower value if you need to find target sequences with more mismatches to your primers. A higher E value should be used if you want more stringent specificity checking (i.e., to identify targets that have more mismatches to the primers, in addition to the perfectly matched targets). On the other hand, a lower E value is recommended if you are only interested in perfect or nearly perfect matches as this will significatly shorten the search time. You can choose to exclude sequences in the selected database from specificity checking if you are not concerned about these. There are a large number of predicted Refseq transcripts in the Refseq mRNA, Refseq RNA and nr database.
Here, we present Simulated Annealing Design using Dimer Likelihood Estimation (SADDLE), an algorithmic framework for designing highly multiplex PCR primer sets. Within this framework, we present an example multiplex primer design algorithm, comprising an algorithm for primer candidate generation and a rapidly computable Loss function for estimating primer dimers. Using the SADDLE, we designed and experimentally tested multiplex primer sets comprising 192 primers (96-plex) and 784 primers (384-plex), and show low primer dimer formation through NGS experiments. Building upon this success, we built a single-tube 60 primer qPCR and Sanger assay to detect and identify 56 gene fusions with clinical actionability for non-small cell lung cancer.
Comments
Post a Comment